Deep Autoencoders
Nonlinear Dimensionality Reduction
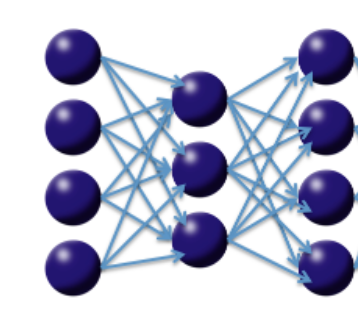
So far we have discussed purely supervised deep learning tasks. However, deep learning can also be used for unsupervised feature learning or, more specifically, nonlinear dimensionality reduction (Hinton et al 2006). Consider the diagram on the following page of a three-layer neural network with one hidden layer. If we treat our input data as labeled with the same input values, then the network is forced to learn the identity via a nonlinear, reduced representation of the original data. Such an algorithm is called a deep autoencoder; these models have been used extensively for unsupervised, layer-wise pre-training of supervised deep learning tasks, but here we consider the autoencoder’s application for discovering anomalies in data.
Use Case: Anomaly Detection
Consider the deep autoencoder model described above. Given enough training data resembling some underlying pattern, the network will train itself to easily learn the identity when confronted with that pattern. However, if some “anomalous” test point not matching the learned pattern arrives, the autoencoder will likely have a high error in reconstructing this data, which indicates it is anomalous data.
We use this framework to develop an anomaly detection demonstration using a deep autoencoder. The dataset is an ECG time series of heartbeats, and the goal is to determine which heartbeats are outliers. The training data (20 “good” heartbeats) and the test data (training data with 3 “bad” heartbeats appended for simplicity) can be downloaded from the H2O GitHub repository for the H2O Deep Learning documentation at http://bit.ly/1yywZzi. Each row represents a single heartbeat. The autoencoder is trained as follows:
1 train_ecg.hex = h2o.uploadFile(h2o_server, path="ecg_train.csv", header=F, sep="\
2 ,", key="train_ecg.hex")
3 test_ecg.hex = h2o.uploadFile(h2o_server, path="ecg_test.csv", header=F, sep=","\
4 , key="test_ecg.hex")
5
6 #Train deep autoencoder learning model on "normal" training data, y ignored
7 anomaly_model = h2o.deeplearning(x=1:210, y=1, train_ecg.hex, activation = "Tanh\
8 ", classification=F, autoencoder=T, hidden = c(50,20,50), l1=1E-4,
9 epochs=100)
10
11 #Compute reconstruction error with the Anomaly detection app (MSE between
12 output layer and input layer)
13 recon_error.hex = h2o.anomaly(test_ecg.hex, anomaly_model)
14
15 #Pull reconstruction error data into R and plot to find outliers (last 3
16 heartbeats)
17 recon_error = as.data.frame(recon_error.hex)
18 recon_error
19 plot.ts(recon_error)
20
21 #Note: Testing = Reconstructing the test dataset
22 test_recon.hex = h2o.predict(anomaly_model, test_ecg.hex)
23 head(test_recon.hex)