Introducing the gemini-cli: Your AI Agent in the Terminal
The gemini-cli as a powerful tool for solo knowledge workers to automate tasks and interact with their digital environment in a more dynamic way.
Note: Some of the following examples using gemini-cli are screenshots (light brown background) and other are the output text copied to this manuscript file for this chapter.
What is gemini-cli and Why It Matters for Solo Workers:
The gemini-cli tool is an open-source AI agent that operates directly from the command line and automates repetitive tasks like working with local files securely, creating digital assets like code and documentation, modifying digital assets, and creating powerful workflows without leaving the terminal.
There is a generous free tier, which is a significant advantage for solo practitioners. The free tier initially uses the best gemini-2.5-pro model but after about ten minutes of session time reverts to gemini-2.5-flash. If you have a Gemini API key, you can pay for all use of gemini-cli and always get the strongest gemini-2.5-pro model.
Getting Started: Installation and Setup:
The gemini-cli command line tool can be installed using either node or npm; here we use npm. You need to insure that an up to date version of npm is installed, then install gemini-client:
1 npm -v
2 npm install -g @google/gemini-cli
As we have seen in previous chapter, you should set an environment variable for your API key:
1 export GOOGLE_API_KEY="YOUR_API_KEY"
You can check that your installation is done correctly by running the gemini command line tool
1 gemini
Note: the first time you run gemini you will be asked to choose a color theme. You will also be asked if you want to login with Google (the free tier), or use the paid tier to always use gemini-2.5-pro:
1 Get started
2 │ How would you like to authenticate for this project? │
3 │ ● Login with Google
4 │ ○ Use Gemini API Key
5 │ ○ Vertex AI
6 │ (Use Enter to select)
If you use the default “Login with Google” a new browser tab will open for you to authenticate gemini-cli. In the user input prompt you can ask general questions or work specifically with the files in the current directory. Here are a few screenshots showing how to use gemini:

As seen in the next figure, you can use the @ in front of a file name to read that file into the work context for the Gemini LLM. If I am using gemini-cli in a directory containing software I often add many source files to the context.
The important thing to remember is gemini-cli can be used in any directory containing digital assets.

Core Concepts and Commands
The interactive prompt is your primary channel for communicating with an AI agent. It’s where you can type natural language questions or issue specific instructions. Commands are special instructions that direct the agent to perform a specific action, rather than just engaging in conversation. These commands are typically prefixed with a special character, like a forward slash (/) or an at symbol (@), to distinguish them from regular text. For example, asking “What is the capital of France?” is a conversational query, while issuing /help is a direct command for the agent to display its help menu. This command-based interaction allows for precise control over the agent’s behavior and access to its various functionalities. You can also use the shell passthrough feature using ! to run regular terminal commands.
Practical Use Cases for the Solo Knowledge Worker:
We will look at three practical examples in this section:
- Writing assistant
- Analyze a CSV data file
- Integrate web search and research in a gemini-cli session
Content creation and writing
The gemini-cli agent tool can be used generally for content creation and repurposing archival material.
As an example, working in the directory for this book manuscript, I can get a summary of a chapter I wrote six months ago:
Analyze a CSV File
I created an empty directory and copied a University of Wisconsin cancer data file labeled_cancer_data.csv to this director, then started gemini-cli:
1 ╭──────────────────────────────────────────────────────────────────────────────╮
2 │ > @labeled_cancer_data.csv look at this file and describe the data columns │
3 ╰──────────────────────────────────────────────────────────────────────────────╯
4
5 ╭───────────────────────────────────────────────────────────────────────────────────╮
6 │ ✔ ReadManyFiles Will attempt to read and concatenate files using patterns: `lab… │
7 │ │
8 │ ### ReadManyFiles Result (Target Dir: │
9 │ `/Users/markwatson/temp/bookex`) │
10 │ │
11 │ Successfully read and concatenated content from **1 file(s)**. │
12 │ │
13 │ **Processed Files:** │
14 │ - `labeled_cancer_data.csv` │
15 ╰───────────────────────────────────────────────────────────────────────────────────╯
16 ✦ This file contains data related to breast cancer diagnosis. Each row represents a
17 sample with the following attributes:
18
19 * Cl.thickness: Clump Thickness (1-10)
20 * Cell.size: Uniformity of Cell Size (1-10)
21 * Cell.shape: Uniformity of Cell Shape (1-10)
22 * Marg.adhesion: Marginal Adhesion (1-10)
23 * Epith.c.size: Single Epithelial Cell Size (1-10)
24 * Bare.nuclei: Bare Nuclei (1-10)
25 * Bl.cromatin: Bland Chromatin (1-10)
26 * Normal.nucleoli: Normal Nucleoli (1-10)
27 * Mitoses: Mitoses (1-10)
28 * Class: The diagnosis class, where 0 typically represents a benign sample and 1
29 represents a malignant one.
30
31 ╭────────────────────────────────────────────────────────────────────────────────╮
32 │ > write a python script to read this CSV file and plot a histogram of clump │
33 │ thickness │
34 ╰────────────────────────────────────────────────────────────────────────────────╯
35 ╭────────────────────────────────────────────────────────────────────────╮
36 │ ✔ WriteFile Writing to plot_histogram.py │
37 │ 1
38 │ 2 import pandas as pd
39 │ 3 import matplotlib.pyplot as plt
40 │ 4
41 │ 5 # This script requires pandas and matplotlib.
42 │ 6 # You can install them with: pip install pandas matplotlib │
43 │ 7
44 │ 8 # Read the CSV file into a pandas DataFrame
45 │ 9 try:
46 │ 10 data = pd.read_csv('labeled_cancer_data.csv')
47 │ 11 except FileNotFoundError:
48 │ 12 print("Error: 'labeled_cancer_data.csv' not found. Make │
49 │ sure the file is in the same directory as the script.") │
50 │ 13 exit()
51 │ 14
52 │ 15 # Check if the 'Cl.thickness' column exists
53 │ 16 if 'Cl.thickness' not in data.columns:
54 │ 17 print("Error: 'Cl.thickness' column not found in the CSV │
55 │ file.")
56 │ 18 exit()
57 │ 19
58 │ 20 # Create a histogram of the 'Cl.thickness' column
59 │ 21 plt.figure(figsize=(10, 6))
60 │ 22 plt.hist(data['Cl.thickness'], bins=10, edgecolor='black', │
61 │ alpha=0.7)
62 │ 23 plt.title('Histogram of Clump Thickness')
63 │ 24 plt.xlabel('Clump Thickness')
64 │ 25 plt.ylabel('Frequency')
65 │ 26 plt.xticks(range(1, 11))
66 │ 27 plt.grid(axis='y', linestyle='--', alpha=0.7)
67 │ 28
68 │ 29 # Save the plot to a file
69 │ 30 plt.savefig('clump_thickness_histogram.png')
70 │ 31
71 │ 32 print("Histogram saved as 'clump_thickness_histogram.png'") │
72 ╰────────────────────────────────────────────────────────────────────────╯
73 ✦ I have created a Python script named plot_histogram.py. You can run it from your
74 terminal to generate the histogram. The script will save the plot as
75 clump_thickness_histogram.png.
76
77 To run the script, use the following command in your terminal:
78 1 python plot_histogram.py
79
80 You will need to have pandas and matplotlib installed. If you don't have them,
81 you can install them with pip:
82
83 1 pip install pandas matplotlib
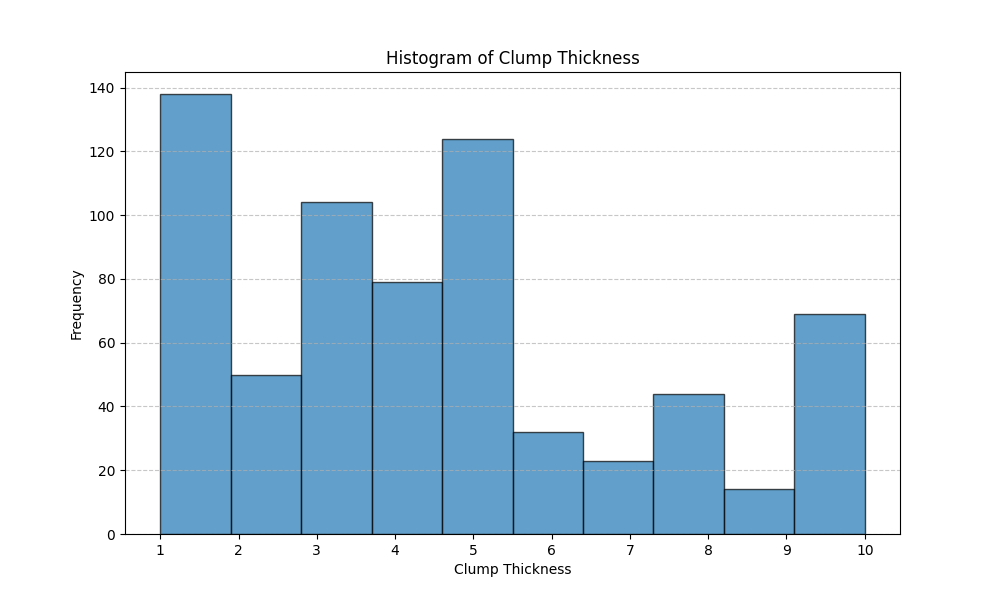
This is routine data analysis that would have taken perhaps ten minutes to perform myself. It took me one minute to analyze the file, run the program gemini-client generated, and view the plot:
Integrate web search and research in a gemini-cli session
You can use Google web search directly in any gemini-cli session as seen here:
